
nrofsubjects: 16;
nrofintervalls: 1;
nrofchannelgroups: 1;
task:2;countforward,countbackward;
color:2;countredsquares,countgreensquares;
between gender:2;male,female;
[0,1,0,1,0,0,1,1,1,1,0,0,1,1,0,0];
EEG and MEG studies are most often analyzed with a special kind of analysis of variance, that accounts also for within subject changes rather than only for between subject differences. EMEGS offers the possibility to calculate this kind of analysis directly, without exporting the data to a separate statistic software package. Moreover, you can choose between a region-of-interest analysis, averaging over sensorgroups and time points, or a complete analysis for every sensor and time point in your data.
To run a repeated measures ANOVA, prepare your data as described:
Each condition for every subject has to be saved as
an SCADS
average file. Every file has to have the same number of points and
number of channels and the same baseline calculation. You need a
textfile, listing the paths of those files on your
machine (a 'batchfile'), with one path per line.This batchfile has
to reflect the design of the planned
analysis, that is, your paths have to be listed according to the
hierarchy of your factors. The lowest level is always the
subject
factor, so you 'll start with one cell for which all subjects
average
files are given.
Beneath that, you list all subjects average files for the next
cell
etc. The subject order has to be identical in every cell, and you
have
to have equal number of subjects in every cell. Please note that
the
structure of the batchfile is identical, wether or not you
have
defined one or more between factor(s)!!!!!
Unequal cell sizes (between) or missing data (within) are
not supported for the matlab-based ANOVA. The R-based ANOVA
however
supports unequal between cell sizes, and the R-based
Mixed-Effect-Models supports both, unequal cell sizes and missing
data.
Cells are ordered from lowest hierarchy position of
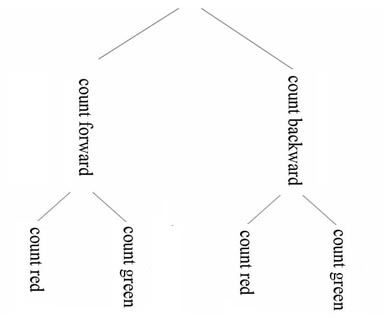
the factor to highest hierarchy position. For instance, consider a
2X2X2 design with the factors 'task'
(count forward vs count backward) , 'color' (count red
squares
vs. count green squares) and 'gender'. Your batchfile for 16
subjects
has to have
the following form:
Once you've prepared this, load one of the listed files in
Emegs2d, set the baseline and display all points. Start Emegs3d.
Here,
load the batchfile as filematrix
(File\FileMatrix\LoadBatchfile).
Then choose \Calculate\Repeated Measures Anova\Define, pick your
preferred
input mode (text or gui (graphical user interface)
),
and enter your
design. The GUI-way is mostly self-explanatory, for the text mode,
you
can follow the example above. With either mode, you always
have
to enter the
following three parts: number of subjects in your sample
(ignoring betweenfactors, that is the total
number subjects in all groups), the number of
channelgroups
and the number
of intervalls using the following syntax:
All other within factor definitions are optional and have to comply with the following syntax:
Between/group factor definitions have the following syntax:
As you can see, one difference to the within factors is the
subject
vector, which specifies the group affiliation of each subject ( in
text-mode, this vector forms a new
line right after the factor declaration ). You can supply any
integer
numbers as group indices, but no strings. These numbers are mapped
to
the cell names according to their value: the lowest number will be
the first group, the next higher number the second group etc. .
Subject vectors have to have one element for each subject, but
subjects
do not have to be grouped according to their group membership (a
vector
like [0 0 1 1] is equivalent
to [0 1 0 1] if your batchfile is ordered accordingly. The second
difference to a within factor definition is the mandatory keyword
'between' before the actual factor definition.
If you whish to calculate the design for all
points and channels and view the output as SCADS average files,
set nrofintervalls to the total number of points
in your data files and nrofchannelgroups to the number of channels
(text) or check the 'all points and channels' checkbutton
(gui). Please note, that with a high number of channels and
timepoints,
this analysis quickly exceeds the available RAM of your computer.
To
effectively
avoid the crash, you can reduce the spatial and temporal
resolution by
using
'Intervall means' and/or 'channelgroups' (see below).
If you want to run one special analysis across a selected number
of
channels and
certain time points, enter the number of channelgroups and the
number
of timeintervalls correspondingly. These values have to match the
channelgroups loaded in Emegs2d and the time/intervall settings in
Emegs3d. If more than one of each is provided, channelgroup and/or
time
will be a separat factor in the ANOVA. Please not that for a true
pointwise
analysis, the whole intervall has to be selected in Emegs3d
including
the baseline points!!!
If you want to use channelgroups and/or intervalls (using
Emegs2d\Calculate\Channelgroups and
Emegs2d\Calculate\Intervall Mean) but still want the output
as continuous SCADS-files, add the line 'continuous results;'
at
the
end of the
design (text) / check the 'continuous results'-checkbox (gui). The
result files will
be stepfunctions of time: all timepoints in a defined intervall
will
have the same value.
This is also true for channelgroups: all sensors in a channelgroup
will
have the identical
stepfuntions. Intervall and channelgroup definition files can
either be
created
and loaded manually or automatically by using the 'Auto
intervalls' or
'Auto groups' menuitems.
The automatic creation usually is much easier and sufficient in
the
case, that you only wish to use
intervalls/channelgroups to reduce memory load.
Click 'OK' and choose \Calculate\Repeated Measures Anova\Run Anova
or
click the
'OK & Run'-button. For a pointwise/continuous analysis, you
will be
prompted
for a target folder, where results are going to be saved. EMEGS
will
save two average file in
SCADS format for every factor and interation in your ANOVA, one
with
p-values, one with the F-Values. For a single analysis results
will be
displayed in new figure, from which you can calculate post-hoc
test,
display means graphically and export the data to a text file.
Post-Hoc Contrasts: Post-Hoc
Contrasts usually are calculated for
significant effects found in an analysis of variance. In EMEGS
this can
be done for one specific analysis of interest and also for every
sensor
and time point. Post-Hocs for one analysis of interest can be
calculated from the output window displaying the ANOVA-results. It
requires that you first start the plotting mode by choosing
'\Graph\Cellplot' in this window and then plot the effect that you
wish
to explore, by adding it's components to the plot on the
anova-plotting
menu (see below). Then you can compare all cells by choosing
'\PostHoc\Entire
family' (without alpha-error correction). Alternatively, you can
enter
specific coefficients for your cells by choosing '\PostHoc\Custom
contrast'. Moreover you can calculate a series of contrasts using
the
Bonferroni-Holm stepdown procedure for alpha-correction by
choosing '\PostHoc\Bonferroni Holm'.
For this, you need to create a contrast text-file, containing your
coefficients for every contrast to calculate with one contrast per
line. EMEGS then calculates these contrasts, sorts them by their
significance level, and tells you which ones can be considered
significant. Results of all the described tests are appended to
the
anova results in the listbox of the results window.
To calculate Post-Hocs for every sensor and time point, choose
\Emegs3d\Calculate\Repeated measures anova\Pointwise post-hocs'. A
window will open, that lets you select an effect or interaction to
be
explored. When you`re done selecting your effect, hit the
'Calculate
post-hocs'-button. EMEGS will then create one average file for all
possible cell comparisons in the selected effect (for the entire
family), containing the uncorrected p-values of the corresponding
contrasts and name the resulting files using the cell labels
specified
in the anova design.
Exporting data to an external
statistics software: To
analyse your data in a methodically more sophisticated fashion,
e.g. to
correct for violations of variance homogenity or other assumptions
or
to calculate certain types of post-hoc contrasts, it is often
necessary
to export your data to a dedicated statistic software. The
ANOVA-plotting module offers a way to export the current data
matrix in
a text-file, that can easily be read by SPSS, Statistica, Jump or
other
packages. To do this, choose \data\export data from the figure
showing
your ANOVA results. You will be asked to choose a decimal
separator and
a filename and filelocation. Variable lables of grouping
factors
are taken directly from your design. Columns for repeated
measurements
however are labelled using the following pattern: 'c' stands for
channelgroup, 't' for time/intervall and 'm' codes the product of
all
other custom within factors. Columns are
ordered according to the hierarchy of the factors. Thus,
the
export for the above example looks something like this:
| subject | gender | m1c1t1 | m2c1t1 | m3c1t1 | m4c1t1 | |
|
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 |
0 1 0 1 0 0 1 1 1 1 0 0 1 1 0 0 |
-3,1144 -9,4074 -1,6262 -2,1788 -1,0859 -2,0606 -1,8005 -2,5449 -2,0271 -1,1013 0,10466 -1,2691 -0,01760 3,4571 0,348 -0,36636 |
-0,51314 2,1428 -5,1445 -6,5715 -5,3422 -5,8692 0,78262 -0,70936 -2,6565 -2,8715 0,013065 0,41345 -1,9967 -1,0716 3,0668 1,6339 |
-1,6344 -2,4501 1,9558 -0,29208 4,8081 1,04 1,8021 1,2636 -3,6415 -3,6158 2,2547 2,2417 3,2311 2,2883 -0,42271 -0,92114 |
2,1613 2,1346 2,6973 0,73483 1,6999 2,8778 3,0983 2,4473 0,17569 0,9375 3,6306 2,697 1,1799 2,5356 6,0943 5,5441 |
As in the example the time factor is limited to 1 intervall and
only
one channelgroup is used, there are only 4 columns corresponding
the
cells of the 'task' and the 'color' factor with 2 gradataions (2*2
= 4
total) each. This arrangement of the data is in line with the way
SPSS
and
Statistica are calculating ANOVAs with repeated measurements and
corresponds the transposed
structure
of the batchfile we used for
calculating the ANOVA.
| |
|
 |
|||
| subject | gender | m1c1t1 |
m2c1t1 | m3c1t1 | m4c1t1 |
| 1 2 3 . . . |
0 1 0 . . . |
-3,1144 -9,4074 -1,6262 . . . |
-0,51314 2,1428 -5,1445 . . . |
-1,6344 -2,4501 1,9558 . . . |
2,1613 2,1346 2,6973 . . . |