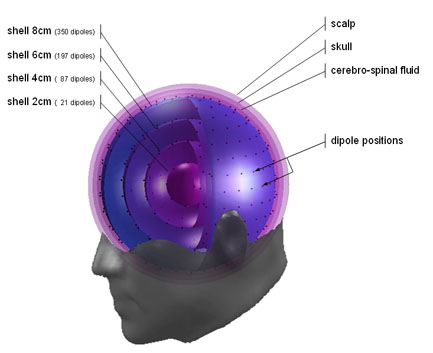
The head model emegs
uses for source localization is sphere model with 4 dipole shells, 1
cerebro-spinal fluid shell, 1 skull shell and the scalp surface.
Dipoles on the shells are equidistantly distributed, resulting in more
dipoles on the outer and less on the inner shells. The picture below
illustrates the model:
Taken seperately, dipoles on each shell can be projected onto a plane
and their absolute strength can be displayed like standard evoked
potentials/magnetic fields in
emegs2d. This is illustrated for the four shells below:
| 8cm
(350 dipoles) |
 |
| 6 cm (197dipoles) |
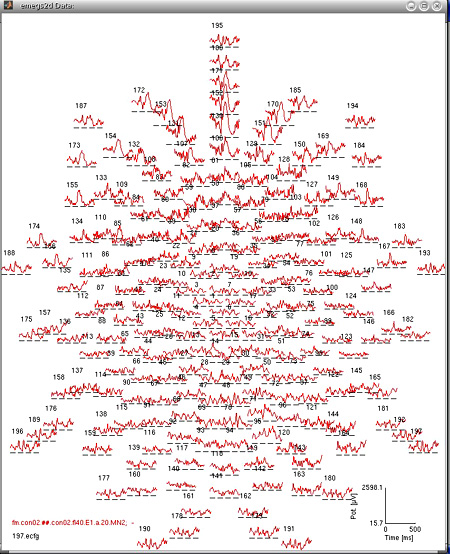 |
| 4cm ( 87 dipoles) |
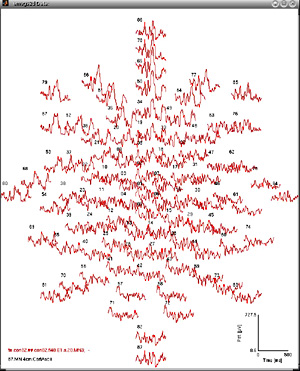 |
| 2cm (21 dipoles) |
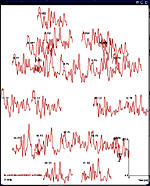 |
Although the inverse solution using the shell model generates 3
dimensional data, analysis of dipole activations has most often been
done for only the third (6cm) shell, as the depth resolution on the
inverse solution is limited (Hauk et. al.???, the preference of
solutions with minimal total power leads to mostly superficial
sources???), and the outermost shell is most prone to noise artefacts.
You can however convert the estimated dipole activation to volumetric
data and browse it with the built-in mri viewer, MRICro or any other
MRI viewer.
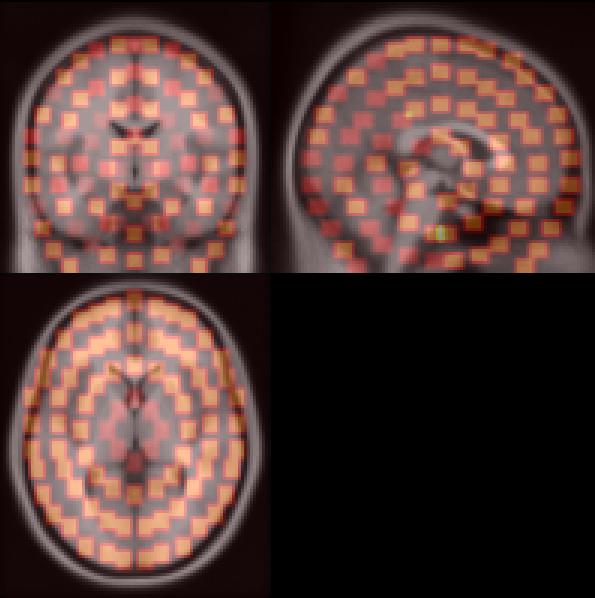
Note that this is a rough approximation, suitable for
exploratory purposes, but hardly for hypothesis testing. The picture
below shows the dipole locations (orange) that are fitted on an
averaged brain. You can save a minimum norm volume by selecting \emegs3d\Calculate\Save MN to Analyse
(91x109).