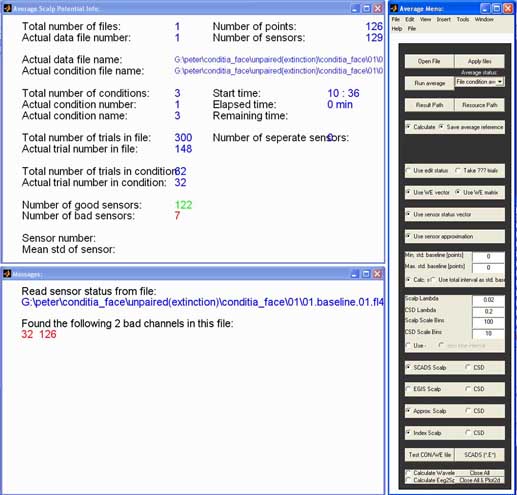
Commands: emegsavg
Based on the thresholds set using EditAEM, EmegsAVG takes the good
trials, approximates missing sensors, and calculates one average
waveform for each condition. The console in action is show below.
In most cases you only need to specifiy 3 options, before running the
average:
(1) first of all you have to choose the data format you're using.
The default 'SCADS *.E1' is shown on a button on upper right of the
panel. Click this button to switch between EGI, Neuroscan, CTF and BTI
format.
(2) choose your segmented data files using the 'Open File'-button in
the upper left corner of the console. You can choose your datafiles one
after the other or open a batchfile. Choose cancel to stop the input
process. Once you've stopped, EmegsAVG will check for the corresponding
editing files (*.AR.WE), the condition-files (*.CON), simply by looking
for files with the same name but different name extensions. If you
accidentially have renamed your files so the segmented data files and
their followers do not match, EmegsAVG will not be able to find them
and run the average. The 'Run Average'-button will be disabled
and EmegsAVG gives you a warning about the missing file in the message
window. EmegsAVG also looks for the default sensor configuration file
for the number of channels in your data and displays its path in the
message window.
(3) Enter your baseline start and end point in the baseline section.
This baseline information will only be used for generating noise
parameters, your data still has to be baseline corrected afterwards.
Now you can start the averaging process, by pushing the 'Run
average'-button. EmegsAVG will display which file it is currently
working on, and how many files are left.
