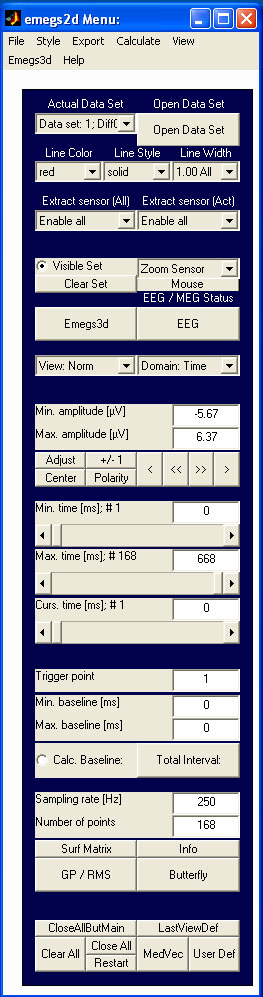
Command: emegs2d
emegs2d is the most powerful tool in the emegs package. It consists of a data window and a control window, with several menus. Its functions are described in the following sections.

Actual Data Set:
Sets the chosen file to actual.
Open Data Set:
Shows
a file open dialog to load a data file to the actual data set position.
LineColor:
Choice of Color of the actual file; if a color map is used please look for Style:Sensor line color in the top menu in this manual
Default for LineColor: 1 red, 2 blue, 3 black and 4 green
LineStyle:
Choice of Linestyle of the actual file
Default for LineStyle is solid.
LineWidth:
Choice of LineWidth of the actual data set (Act) or all data sets (All)
Settings
will be used by Global Power
Plot, Butterfly Plot and Zoom Sensor Plot
Visible:
Off sets all data sets of chosen sensor invisible. Press once more to make them visible. Invisible sensors will be extracted of all further analysis (Global Power, Surfaces, Stats etc.)
Visible Act:
Off sets only actual set chosen sensor invisible. Press once more to make them visible.
Invisible sensors will be extracted of all further analysis (Global Power, Surfaces, Stats etc.)
Visible Set:
Off sets all sensors of the actual data set invisible. Press once more to make them visible.
Clear Set:
Clears the actual data set (cell) (does not delete not the original file but only the imported data matrix).
Zoom Sensor:
Mouse (is only possible in Matlab4 because of a bug in ginput.m using Matlab5)
After choice of Zoom channel use the mouse to zoom special channels. This channel will be shown in an extra window.
Channel Name: Choose the channel you want to zoom. This channel will be shown in an extra window.
Emegs3d:
Launches the Emegs3d (Emegs3d.m) part of the program containing the features:
Scalp potential/magnetic field interpolation
Laplacian of scalp potential/magnetic field interpolation
Intracranial scalp potential/magnetic field estimation
L2-Minimum-Norm estimations
Export of batch file calculation. (Sensors, Regions, Dipole Indices, time points or time intervals of interest. This Manual is under construction.
EEG/MEG Status:
Changes EEG/MEG Status to either EEG or MEG
View:
Top: View from top at elctrodes which Z-value is bigger than 0 (nose to top).
Left: View from left (nose to left) at electrodes which X-value is smaller than 0.
Right: View from right (nose to right) at electrodes which X-value is bigger than 0.
Front: View from front (right ear to left) at electrodes which Y-value is bigger than 0.
Back: View from back (right ear to right) at electrodes which Y-value is smaller than 0.
Norm (default): All electrodes are visible.
It is possible to set the positions of the time axes directly:
Choose To View in View menu
Choose the menu: File:Electrode File Format:Cart. Ascii (set the electrode format to ascii)
Choose the menu: File:Open Electrode File
and choose the file ...:Emegs2d3d:Emegs2dUtil:Box129CartAscii.ecfg
Look at this file (Box129CartAscii.ecfg ) to understand the format:
First column : x-axes runs from 1 to 10
Second column: y-axes runs from -1 to -14 (because the channel with biggest y-value is set to top)
The third column does not matter (but has to be set to a value) because the view is set to Top.
In this way every user is able to create his own axis positions.
Domain:
Time (default): Shows data in time domain
Std: Shows the given data of standard deviation (if this info is not given all std values should be zero)
Power FFT: Shows power spectra of data in frequency domain
Amp. FFT: Shows amplitudes of spectra in frequency domain (sqrt(real^2+imag^2))
Phase FFT: Shows phase of spectra in frequency domain (arctan(real/imag))
PCA: Manual needs to be written
Wavelet Ampitude: Manual needs to be written
Wavelet Phase: Manual needs to be written
Min. Amp; Max. Amp:
Defines the visible interval of amplitudes
Adjust:
Adjusts visible interval of amplitudes to minimum and maximum values across all visible data.
Center:
Centers the visible interval of amplitudes
+/- 1:
Shows and changes the fast amplitude change value:
+ :
Increases the given maximum with the given fast amplitude change value
- :
Decreases the given minimum with the given fast amplitude change value
+/- :
Both
View:
Inverts the view: Negative Up or Negative Down.
<:
Decrease visible minimum or/and maximum (depending on +/-1)
>:
Increase visible maximum or/and minimum (depending on +/-1)
<<:
Decrease amplitude change value using a factor of 10
>>:
Increase amplitude change value using a factor of 10
Min. Time; Max. Time:
Defines the visible interval in time. Choose the time interval in ms from the given trigger point. Example: Data matrix with 500 points in time and a sampling rate of 250: The trigger occurs at the 100th point counting from the beginning of file.
If the user would set Min. Time to zero, the first visible point of time is the trigger point. If the user would set Min. Time to -100, the first visible point of time is the 75th point of the data matrix. The time in ms are rounded to the next possible data point. In this example 1 or 1.9 would be rounded to zero, 2 and 3.1 would be rounded to 4 ms.
TriggerPoint:
Defines the trigger time. This point has to be given in absolute number of points counting from the beginning of the file. (not in ms)
Cursor Time:
Defines the cursor time. This time has to be given in ms from the trigger time. (compare with Min. Max Time)
Min. Baseline, Max. Baseline:
Defines the interval used for baseline correction. This time has to be given in ms from the trigger time. (compare with Min. Max Time)
If Min. or Max. Baseline are set: This does not mean that the baseline correction is done.
Calc. Baseline radio button:
Switch the radio button of baseline correction on to do the calculation. Switch the radio button off to get the original uncorrected data
Total Interval (shortcut):
Calculates baseline across whole time interval
Sampling Rate:
look at: Calculate:Sampling
Number of points:
Shows the number of points of the actual data set.
This could change if using down- or up- sampling in the Calculate:Sampling top menu
GP/RMS:
Calculates Global Power of visible files (cells) and plots them in an extra window
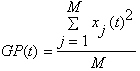
or Root Mean Square
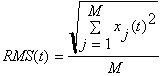
M is the number of sensors
![]() is the electrical potential or
magnetic field of the sensor j at the point of time t.
is the electrical potential or
magnetic field of the sensor j at the point of time t.
Use the lowest menu button in this window to switch between RMS or GP.
The line style is identical with the line style in the main window.
The trigger time point is identical with the trigger time point in the main window.
The baseline correction t is identical with the main window baseline correction.
The user could use Std weight to use the Std data to weight the channels while calculating the Global Power
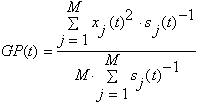
or the Root Mean Square
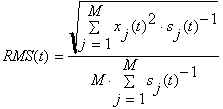 .
.
If no Std data is available they are all set to 1. Consequently a weighting does not alter the data.
Butterfly:
Plots all visible channels of the main window in one extra window. Invisible time courses in the main window are also invisible in this extra window.
Linestyle, time intervals, baseline correction amplitudes etc. are identical with the given data in the main window.
Surf Matrix:
Surfs the Main Window channels in an extra Surf Data Matrix Window. Each Cell (File) will be surfed in a special subplot
Info:
Gives information about linestyle and names of all files (and/or number of cells).
Clear All:
Clears all given data.
Close All:
Closes all open Matlab windows except Command window.
Attention!! Closes also windows of different Matlab programs e.g. SPM
XCorrChan:
For your own purpose. Editing in Emegs2d
MedVec (SCADS only) :
Plots information about single trial medians calculated in AvgApprox.m
File:Open Data Set
Default: If Default File format is not given the file will revert to EGIS average format.
Opens FileSearchWindow with FileMask
*.ave* if averaged EGIS file
*.gave* if grand averaged EGIS file
*.ses* or *.E* if EGIS session file
* if Ascii
* if Ascii with Header; Header: Number of Channels, Number of Points
*.at* if SCADS Format
*.avr* if Ascii Besa Format
*.raw* if MEG ascii file
Reads the given data matrix of file and displays it in Actual Data (highest left button).
Open Data Set 1 Opens first data set
Shortcut: Apple (Control on PC or Unix) plus h.
Open Data Set 2 Opens second data set
Shortcut: Apple (Control on PC or Unix) plus j.
Open Data Set 3 Opens third data set
Shortcut: Apple (Control on PC or Unix) plus k.
Open Data Set 4 Opens fourth data set
Shortcut: Apple (Control on PC or Unix) plus l.
Open Data Set i+/-j set (SCADS format only) Asks for first file name with extension (*.at(i)*) and opens maximum additional 3 files (*.at(i+/-j)*)) as long as they exist in the identical directory.
Shortcut: Apple (Control on PC or Unix) plus m.
Open Batch Set (SCADS format only) asks for a (*.at*) batch file containing full file names (including path) of maximum 4 (*.at*) existing SCADS files to.
Shortcut: Apple (Control on PC or Unix) plus ,.
Important
If using MEG data: Overlaying time courses of different data sets need identical sensor and head model configurations. Therefore data sets with set values higher than 1 (Data set 2; Data set 3 and Data set 4) use the sensor and head model configuration of Data set 1. If the user reads a new Data set 1 the program automatically searches for the new corresponding Sensor- and Center of Sphere configuration.
If a data file is opened without a given sensor configuration file the program searches for file
...:Emegs2d3d:Emegs2dUtil:SensorCfg:Number of Channels.ecfg Sensor configuration if SCADS Sensor configuration format.
or
with sensor configuration with the same name as the data file and in the same folder but with extension .pmg instead of .raw if the sensor format is MEG on sphere or MEG no sphere
See Open Sensor Set for other sensor configuration formats
If the sensor configuration file does not exist Open Sensor File will be activated
The program will test whether the number of channels in the data file equals the number of channels in the sensor configuration.
If no an error message will be displayed -
This data file contains Nchan channels
The sensor configuration file contains Nchan channels
Please choose a different data file or a different sensor configuration.
File:Open Sensor Set
Default: If not nominated the default configuration is -
SCADS format if EEG data
MEG no sphere format if MEG data
If the data file is already chosen program searches for the sensor configuration file with corresponding FileMask.
Opens FileSearchWindow with FileMask:
*.int2str(NChan).elp if Besa sensor configuration
*int2str(NChan).ecfg if Egis sensor configuration(Eugene electrode configuration is not yet given)
*int2str(NChan).ecfg if sensor configuration format is SCADS
* int2str(NChan).CartAscii if sensor configuration format is CartAscii
* int2str(NChan).SpherAscii if sensor configuration format is SpherAscii
* int2str(NChan).AxesAscii if sensor configuration format is AxesAscii
searches for the sensor configuration with the same name as the data file and in the same folder but with extension .pmg instead of .raw if the sensor configuration format is MEG on sphere or MEG no sphere. If this file does not exist a FileSearchWindow will be opened with the FileMask:
*.pmg
If no data file is chosen the command window displays options for nominating file with sensor configuration. The FileMask for the type of data must be nominated in the Dateityp window. e.g if SCADS format is used the sensor configuration may be nominated by searching for the file named ...:Emegs2d3d:Emegs2dUtil:SensorCfg... the options available for sensor configuration will be displayed.
Opens FileSearchWindow with FileMask:
* elp if sensor configuration format is Besa
* ecfg if sensor configuration format is SCADS
* CartAscii.ecfg if sensor configuration format is CartAscii
* SpherAscii.ecfg if sensor configuration format is SpherAscii
* AxesAscii.ecfg if sensor configuration format is AxesAscii
*.pmg if the sensor format is MEG on sphere or MEG no sphere.
If the sensor locations are not positioned onto a sphere the user will be asked whether this is required.
If a sensor weighting file is available it has to be of the Header Ascii Matrix form.
These values are used to weight the sensor positions in the best sphere fit calculation.
If no weighting file is used all sensor positions are weighted with a value of 1.
CSD or Intracranial mapping of scalp potentials must be represented in the spherical form as calculations for these are limited to the spherical form. However, it is possible to calculate Minimum Norm with this configuration.
This is especially of importance if the sensor configuration is of MEG type.
In case of MEG sensor configuration:
MEG on sphere - the sensor positions will automatically be projected onto a sphere. Now interpolated field maps, laplacians and intracranial field estimations are possible since these methods require spherical models.
The calculation of Minimum Norm uses the original not projected sensor configuration.
MEG no sphere - these methods are not available. However, the calculation of Minimum Norm using this original not projected sensor configuration is more reliable than the projected one.
In the next step the program searches for the MEG center of spherical head model information with the same name as the data file and in the same folder but with extension .cot instead of .raw if the sensor configuration format is MEG on sphere or MEG no sphere.
If such a file does not exist is not possible to correct the MEG sensor positions. The center of sphere will be assumed to the origin [0 0 0].
File: Data Format
This
command allows the type of data
collected to be nominated and searches for the appropriate data under
the following FileMasks. If
Data Format is AUTO FileMask is *.*.
Data Set Format will be chosen automatically depending on the files
extension.

NeuroScan Average
Opens FileSearchWindow with FileMask *.avg* (ReadNeuroscanStruct.m)
NeuroScan Continous
Opens FileSearchWindow with FileMask *.cnt* (ReadNeuroscanStruct.m)
Manual needs to be written
SCADS Wavelet Amplitude
file (Result of SCADS WaveApp.m based on SCADS .app files, results of AvgApprox.m)
Opens FileSearchWindow with FileMask *.awa*
SCADS Wavelet Phase
file (Result of SCADS WaveApp.m based on SCADS .app files, results of AvgApprox.m)
Opens FileSearchWindow with FileMask *.pwa*
EGIS Average file
Opens FileSearchWindow with FileMask *.ave*
Now the ActualData button (highest left button in MainWindow) becomes the first cell and first subject and OpenData (Averaged file) menu (highest right button in MainWindow) will show all possible cells and subjects of this data file. Now the user can choose any of these cells and subjects for a given ActualData number or choose a new file using the New File choice given in the OpenData (Averaged file) menu .
ð Starts again File:Data format.
Egis Grand Average
Opens FileSearchWindow with FileMask *.gave*
EGIS Session file
Opens FileSearchWindow with FileMask *.E*
Now the ActualData (highest left button in MainWindow) becomes the first Cell and first subject and OpenData menu (highest right button in MainWindow) will show all possible cells and trials of this data file. Now the user can choose any of this cells and trials for a given ActualData file number or choose a new file using the New File choice given in the OpenData menu .
=> Starts again: File:Data format
SCADS
Opens FileSearchWindow with FileMask *.at*
Ascii Matrix
Opens FileSearchWindow with FileMask *
Opens a special window named Import Ascii File Information.
This window shows the number of all read data points = NCount.
Number of sensors is set to default 129
Number of conditions is set to default 1
The number of points per condition will be calculated. If this number is odd => Error message. The user has to change the number of sensors or conditions until (Number of sensors*Number of Channels*Number of Conditions)=NCount
The Conditions-Channels-Time menu defines the ordering of Condition, Channel and Points given in the file.
If Number of Channels*Number of Points.*Number of Conditions=NCount
the user is allowed to choose one of these data sets and this data will be set to the given ActualData number in the MainWindow (top left button)
Ascii Header Matrix File
In this case the first ascii value in the ascii file defines the number of channels and the second the number of points.
The following (Number of Channels*Number of Points) value becomes the new data matrix .
If Ncount (number of all read data points) > (Number of Channels*Number of points)à Error message
If Ncount (number of all read data points) > (Number of Channels*Number of points)+2à Warning
Float32 Matrix
As with Ascii Matrix but Float32 binary data
Float32 HeaderMatrix
As with Ascii Header Matrix but Float32 binary data
Ascii Besa
Opens FileSearchWindow with FileMask *.avr*
MEG BTI Raw Ascii
Opens FileSearchWindow with FileMask *.raw
(If any data file is imported without information about the sampling rate (i.e. all imported matrices) the sampling rate value in the Emegs2d menu window will be set automatically to 1. Change this value to the correct sampling rate if known by typing the value in the sampling rate edit text menu (Emegs2d menu figure)
File: Sensor Format
This command allows the sensor configuration to be selected. The following types of sensor configuration are possibilities with this command.
Auto Axes
If no sensor location information is available or for a brief overview of the data choose this Sensor Format option.
The Emegs2d Data figure will automatically be divided in a number of rows and columns showing all time courses using these axes. The use of surface methods (Emegs3d) are not possible without information about the sensor locations.
MEG no sphere
.pmg ascii file from BTI
MEG on sphere
Opens a .pmg ascii file and calculates the best fitting sphere for the sensor info. Given (if needed) and then projects sensor position onto this sphere.
BESA EEG
Number of sensors.elp filemask ascii file
EGIS
Not available at this stage
SCADS
No of channels. Ecfg binary file (SaveEcfg.m)
Cartesian Ascii
Header ascii file filemask cartascii.ecfg. No. of channels initial no. and number of rows second no. (cartesian format uses 3 x,y,z)
Spherical Ascii
Sperascii.ecfg filemask. Header ascii file 2 possibilities 1 3 values, theta polar angle, phi azimutal angle, radius(defaults to 1 if radius not given). Sphere of best fit is calculated and sensor position projected onto this sphere if radius is given.
Axes Ascii
To define the sensor axes positions directly choose: Axes Ascii
The View will be set automatically to Top.
x-position (left-right): Normalize axes positions between minimum and maximum of the first row of given ascii text file.
y-position (bottom-top): Normalize axes positions between minimum and maximum of the second row of given ascii text file.
Example: 129AxesAscii. sensor configuration.
The use of surface methods (Emegs3d) are not possible without information about the sensor locations.
10/20 System
To choose electrode positions of the international 10/20 System select this menu.
10/10 System
To choose electrode positions of the international 10/20 System select this menu.
An extra window will be opened and the command given to select the order of channels in the data based on an assumed spherical head and standard 10/20 system placement (Terrence Lagerlund et al. 1992) or Besa standard 10/10 system respectively.
The channel order has to coincide with the order in the data file, do not choose channels randomly.
After ordering the channels correctly select Apply. The electrode file will be saved in SCADS binary, cartesian ascii and spherical ascii format (first filename needs to be nominated)
To change special positions that are not standard 10/20 or 10/10 positions edit the ascii files using an editor eg. word.
Use the Default file name NChan.ecfg for SCADS format to use this as default.
For further details on sensor file formats
See File:Open Sensor File
File : Default File Auto
Automatically
sets nominated default
values for individual files
If
this feature is highlighted the program
searches for a file with the name DataFolderName.udef when a file is
first opened. While DataFolderName is the name of the actual data file
containing folder. If such a file does not exist the user is asked to
open a file with the FileMask *.udef containing the wished View Default
informations; Press escape if you dont want to use any view default
file.
File: Default File: Open
Opens the FileSearchWindow with FileMask *.udef
Loads all given default information
See
File: Default File: Save
File: Default File: Save
Stores all set values of
channels, time , amplitudes and style information
in a file which has the FileMask *.udef
opened with the auto function when data next used with filemask.udef
File: Default File: Info
Shows
the name of the View default file
which is in use.
Style : Axes Width
Choose relative width of single sensor axes
Style : Axes Height
Choose relative height of single sensor axes
Style : Sensor line color
Using this feature its possible to display different sensors or sensor groups with different colors.
Flip Color Map flips the colormap if Line Color is a colormap e.g. hsv, hot, cool.
The colors are chosen based on the color map given in the Emegs2d main menu windowLine Color (hsv, hot, cool etc.) .
The values used are:
Use default vector => channel number
Use number of trials vector => number of averaged trials
Use Std vector => the mean standard deviation (if given)
Use Channel Groups => channel groups (if given) colormap set automatically jet and Line-Width automatically 3 Act in order to visualize channel groups
Use LineColor File
The assigned numerical values for different channels corresponding to colors on the color map could be imported using an ascii file in matrix format from the LineColor file (follow path ..Emegs2dUtil: LineColor: options displayed [e.g 12Groups, VisualStdLineColor format]).
This option could be used to assign colors to groups of sensors or to highlight areas of particular interest.
Style : Text
The plot labels and additional information to be displayed may be chosen with this option.
To change more than one label choose the menu again and set the desired option.
(If Standard Deviation values are not given they will be set to 1).
(The color of Cursor Amplitudes is identical to the Actual File color).
Style : Zeroline
Display on/off and style (color and line style) of zeroline displayed
Style : Triggerline
As above for Triggerline
Style : Cursorline
As above for Cursorline
Style : X-tick
Time intervals displayed on x-axis.
Style : Background Color
Choice
of white or black.
Export: DataFile
This function allows the Actual Data Set to be exported in a variety of formats
E.g Export: DataFile: SCADS format
Export: DataFile :SCADS format (for 10/20 format data)
Export: DataFile :Ascii Header Matrix
Export: DataFile :Ascii Besa
Export: Datafile :Egis (not currently
available)
Export: SensorFile
Function allows the exportation of the sensor configuration in different formats:
E.g Export: MEG BTI format
Export: SensorFile :Besa (not currently available)
Export: SensorFile: Egis (not currently available)
Export: SensorFile :SCADS format
Export: SensorFile :Cart. Ascii format
Export:
SensorFile :Spher Ascii format
Export:
GP / RMS
To export GP / RMS values these values have first to be calculated using GP/RMS button in the MainWindowMenu
Export: GP / RMS :All Points
The calculated values will be exported to an ascii file. File name has to be given by user.
Export: GP / RMS :Cursor Points
GP/RMS Cursor exports only the cursor indicated on the x axis values to an ascii file.
Export: Userdata
Exports the least calculated values to either an ascii or a besa file.
Export: Userdata Info
States
the nature of Userdata.
Calculate
:Global Dissimilarity
Actual data set or Dataset 1 & 2 calculates the global dissimilarity
(Lehmann et al., 1987) for susequent time points (actual dataset) or
for corresponding time points (Dataset 1 &2) and plots them in a
separate figure. It is calculated by normalizing the signal on all
channels by the square root of it's global power (it's Root Mean
Square) and then averaging the squared differences between
corresponding channels of subsequent points (Actual data set) or two
loaded data sets (Dataset 1&2). It ranges between 0 (maximal
homogenity) to 2 (maximal dissimilarity), and describes topographical
properties independent of absolute amplitudes.
Calculate :Minimum Cluster
Size
Applies a spatio-temporal
correction, that can be used as an alternative of alpha-correction for
multiple testing. It operates on a dataset containing p-values and sets
all points to zero (non-significant), that do not have a user entered
number of significant channel neighbour points and do not have a user
entered number of preceding or following significant time points. The
definition of what is considered a channel neigbour is determined
by a maximum spherical distance in degrees, that can lie between two
sensors. Thus a larger maximum distance will result in less
conservative correction. Maximum spherical distance, minimum number of
channel neighbours and minimum number of points in a row must be
entered before running the corrrection by calling the menu item
\Calculate\Min.Cluster Size\Set Size.
Calculate :Gradients
All data sets or actual data set calculates the temporal gradient (gradient.m; difference from point to point) for all or just for the actual (given in actual data set menu) data set.
Calculate :Dipole Density (in progress; manual needs to be written)
Calculate :t-test (just SCADS format; under construction)
Asks for a batch file matrix containing average SCADS files (*.at*).
Paired: tests pairs of files (1 vs. 2; 3 vs .4, etc.)
Symmetry: tests for anterior-posterior or left-right asymmetry.
Absolute takes the absolute value (just positive; abs(data)) of the original data (positive and negative)
Calculate :Extreme Values
Calculates for each sensor absolute, relative or absolute of relative minimum, maximum or both in the chosen time/frequency/etc. interval.
Calculate :Absolute: The absolute maximum, minimum within the given interval. Absolute extreme values may be interval borders.
Calculate :Relative: All relative maxima, minima within a chosen interval. This calculation checks for a zero gradient. Thus, an interval border can not be a relative extreme value.
Calculate :Absolute of Relative: The absolute maximum, minimum across all relative maxima, minima.
Open identical files in actual file slot 1 and 2 and use one of the features for actual files as an example.
Using features of absolute extreme values the user may export extreme values and extreme indices (points) using the Export User Data function. Please acknowledge that indices are given in points (~time, ~frequency ...) starting with the minimum interval point.
In case of maximum or minimum the first row writes extreme values, the second extreme indices for all sensors. In case of both four rows are exported. The first two rows are reserved for the maximum the third and fourth are used by the minimum values and indices.
Use Export Data File function in order to export relative extreme values and indices.
Calculate : Wavelet (calls WaveApp.m)
Manual needs to be written
Calculate : X-Correlation
Manual needs to be written
Calculate :ProMax
Manual needs to be written
Calculate :VariMax
Manual needs to be written
Calculate : ICA
Manual needs to be written
Calculate :PCA
All data sets or actual data set calculates PCA for all or just for the actual (given in actual data set menu) data set.
Original data set calculates PCA based on the original loaded data set (independent of the currently visible data set)
Actual data set calculates PCA based on the currently visible data (and therefore based on the currently given time, frequency or PCA component interval)
Data matrix calculates PCA based on the given data matrix
Covariance matrix calculates PCA based on covariance matrix of the given data matrix
Correlation matrix calculates PCA based on correlation matrix of the given data matrix
(with choose of Domain: PCA (domain menu) the PCA of all original data sets will be calculated)
The PCA uses the Matlab program SVD:
[U,S,V] = SVD(X) produces a diagonal matrix S, of the same dimension as X and with nonnegative diagonal elements in decreasing order, and unitary matrices U and V so that X = U*S*V'.
The program calculates the variance vector (v) in the following form:
v = (diag(S)).^2/sum(diag(S).^2)
An additional figure widow will be opened. The left subplot shows the whole variance vector. The right one just the elements which are bigger than 0.01 (if all elements are bigger than 0.01 just one plot will be shown). Color and style of lines corresponds to the defined ones in the Emegs2d main menu.
A second additional figure window will be opened displaying just the time courses (row vectors) of the matrix V which corresponding elements of the variance vector are bigger than 0.01. Color and style of lines corresponds to the defined ones in the Emegs2d main menu.
If the given matrix X is of data matrix form (not of covariance or correlation matrix form) the matrix U will be shown in the Emegs2d Data Figure changing the given domain to PCA. Color and style of lines corresponds to the defined ones in the Emegs2d main menu. The minimum point will be set to its first component (the first row elements (channels)) In the same way trigger and cursor point will also be set to the first component. Since the number of rows of matrix U is equal the number of channels and the number of columns corresponds to the maximum number of PCA components (minimum of number of channels and given number of points (depending on the number of chosen points of visible interval (not the length of the interval in ms or Hz))) all data points corresponding to not existing components (components higher than the maximum number of PCA components) are set to zero (in future versions it will not be possible to choose such components).
The
program calculates the significance
levels (95% and 99%) for the matrix U based on the maximum number of
PCA components and signs all channels which values of the given cursor
component are higher (significant) than theses levels (95%=>* and
99%=>**). The color of the stars corresponds to the defined colors
in the Emegs2d main menu.
Calculate :Polyfit
The Polyfit function finds the coefficients of a polynomial p(x) of the given degree n and fits the data p(x(i)) to y(I) in a least square sense. The result p is a row vector of length n+1 containing the polynomial coefficients in descending powers.
[i.e p(x)=p1x^n+ p2x^(n-1)+ p3x^(n-2)++pn+1];
The polyfit will be calculated using just the visible time interval (in the Emegs2d Data figure) (not all time points). The whole data set must be made visible to use all values.
It is possible to export the polyfit data matrix or the vector of the calculated polynomial coefficients of each sensor using the Export UserData option (see Export; UserData).
At least the difference of the given data set and its polyfit will be plotted.
In this way it is possible to correct drift artifacts using a degree of 1 (detrending).
Warning:
Detrending should only be used on
the base of the baseline interval. Care must be taken using the
whole epoch because the detrending could at least be based on real
signal with a least square deviation different from zero. (e.g in the
case of a large P300)
Calculate :Channel Groups
Open group file: Load a group matrix file with FileMask .Ascii (matrix file format) with first line the number of group and second line the sensor number. (See file 12Groups in Emegs2dUtil:Groups as an example).
Mean: Calculates the mean over given groups of the given time interval.
Use Export:User data to export this calculated data to file.
Sum: Calculates the sum over given groups of the given time interval.
Use Export:User data to export this calculated data to file.
Calculate :Egis session file:View Egis data
Calculate :Egis session file:Edit Egis data
if data format is an Egis session file this feature is enabled.
To scroll through the data trial- or view epochs use:View Egis data.
An extra window will be opened containing the following buttons:
Given: Shows the given Edit Status
red if bad
green if good
blue if questionable
white if not edited
Default:
(for future use; if an external edit vector is given)
>: Scrolls to the next
good trial (if the Next Good radio button is enabled)
bad trial (if the Next Bad radio button is enabled)
questionable trial (if the Next ??? radio button is enabled)
<: Scrolls to the last
good trial (if the Next Good radio button is enabled)
bad trial (if the Next Bad radio button is enabled)
questionable trial (if the Next ??? radio button is enabled)
>>: Scrolls to the first
good trial (if the Next Good radio button is enabled)
bad trial (if the Next Bad radio button is enabled)
questionable trial (if the Next ??? radio button is enabled)
of the next cell.
<<: Scrolls to the first
good trial (if the Next Good radio button is enabled)
bad trial (if the Next Bad radio button is enabled)
questionable trial (if the Next ??? radio button is enabled)
of the last cell.
To edit the data by trial groups manually use:Edit Egis data
Buttons:
As with the View Egis data option with the additional buttons:
Good Trial sets the Edit value of this trial good
Bad Trial sets the Edit value of this trial bad
??? Trial sets the Edit value of this trial questionable
if the Auto Next radio button is enabled the
next trial (if the Next Good or the Next Bad or Next ??? radio button is not enabled)
next good trial (if the Next Good radio button is enabled)
next bad trial (if the Next Bad radio button is enabled)
next questionable trial (if the Next ??? radio button is enabled)
will be shown following the actual trial edition.
Calculate :Your Own: Actual Data Set
Opens the file ...Emegs2d3d:Emegs2dUtil: CalcUserFile.m
and does the given calculations just for the given actual file (cell)
If this calculation is not possible =>Error message
Calculate :Your Own: All Data Sets
Opens the file ... Emegs2d3d:Emegs2dUtil: CalcUserFile.m
and does the given calculations for all given files (cell)
If this calculation is not possible =>Error message
Reset Calculate: Your Own: Reset Actual Files
Sets Actual data back to the Actual data which was given before the calculation carried out.
Calculate :Your Own: Reset All Files
Sets All data back to original status.
Calculate :Average:(Data set 1, Data set 2) => 3
Calculates the average of data set 1 and data set 2 and sets the result to actual file number 3.
Calculate :Average:(Data set 1, Data set 2) => 4
Calculates the average of data set 1 and data set 2 and sets the result to actual file number 4.
Calculate :Average: Use Standard Weight Matrix: (Data set 1, Data set 2) => 3
Calculates the average of data set 1 and data set 2 weighting each of the data points by using the given standard values and sets the result to actual file number 3.If no standard values are given all standard values are set to 1 and consequently the weighting is meaningless.
Calculate :Average: Use Standard Weight Matrix: (Data set 1, Data set 2) => 4
Calculates the average of data set 1 and data set 2 weighting each of the data points by using the given standard values and sets the result to actual file number 4.If no standard values are given all standard values are set to 1 and consequently the weighting is meaningless.
Calculate :Difference:Data Set 1- Data set 2 => Data set 3
Calculates the difference between Data Set 1 and Data Set 2 (Data set 1 Data set 2) and sets the result to actual file number 3.
Calculate :Difference:Data Set 1- Data set 2 => Data set 4
Calculates the difference between Data Set 1 and Data Set 2 (Data set 1 Data set 2) and sets the result to actual file number 4
Calculate :Difference:Data Set 2- Data set 1 => Data set 3
Calculates the difference between Data Set 2 and Data Set 1 (Data set 2 Data set 1) and sets the result to actual file number 3.
Calculate :Difference:Data Set 2- Data set 1 => Data set 4
Calculates the difference between Data Set 2 and Data Set 1 (Data set 2 Data set 1) and sets the result to actual file number 4.
Calculate :Absolute
Calculate :Absolute: Actual Data Sets
Calculates the absolute values of the actual file
Calculate :Absolute: All Data Sets
Calculates the absolute values of all files
Calculate :Filter
If a data set is available it is possible to
Calculate :Filter: Set filter file
set filter for high and/or lowpass filtering
Calculate :Filter: Save filter file
save filter coefficients in the file with filemask* int2str(NChan).hf for highpass or *int2str(NChan).lf for lowpass coefficients
Calculate :Filter: Open filter file
open filter coefficients in file with filemask *int2str(NChan).hf for highpass or *int2str(NChan).lf for lowpass coefficients
Calculate :Filter: All files or Actual file.
filter the all or jsut the actual data set using the set or read filter coefficients
Calculate: Invert
Calculate :Invert : All Data Sets
Invert (Multiplication by -1) the values of All Data Sets
Calculate :Invert: Actual Data Set
Invert the values in the Actual Data Set
Calculate :Normalize
Normalizes Actual Data Sets or All Data Sets in different ways:
Between -1 and +1
Between 0 and +1
With std=1 and mean=0 (z-transformation)
or sets values
Absolute maximum of each data set to 1
Median across all absolute values of each data set to 1
Mean across all absolute values of each data set to 1
Calculate :Potential
Calculates integral or mean for each channel and each file and shows the results in the command window.
The Integral is given in µV x ms. ( the sum of all amplitudes in the chosen time interval multiplied by the sampling time (1000/SamplingRate) )
The Mean is given in µV ( the sum of all amplitudes in the chosen time interval divided by the number of time points in this interval)
Transfer this data into text or statistics programs by:
1. Using Export:User data and exporting these values to a text file.
2. Copy the values directly from the command window and paste it into a text or statistics program.
Calculate: Potential: Integral or Mean: Interactive
It is possible to choose one or more time intervals interactively in the command window
or by using a given file
Calculate :Potential:Integral or Mean:File
In this case the given time intervals in the file ...: Emegs2d3d:Emegs2dUtil: Intervals:IntegralInterval.txt will be used.
Calculate :Potential:Integral or Mean:Default
uses the last given intervals
Important:
The time values have to be given in time points counting from the begining of data set and not in ms counting from trigger. The intervalls may be different from the intervall which is given in the main window.
Calculate : Rereference
Rereference all files (or Cells) (using All files) or just the working file (using Actual file) using a different reference channel. The program shows the time course of the new reference in an extra window.
If the user chooses Special, a new window will be opened in which the new reference channel or channels can be chosen. If the user chooses more than one channel the average of these two channels are used. (Linked ears uses the average of both mastoids)
Calculate: Sampling
If enable is on a change of the Sampling rate value in the main edit window results in a calculation of the given data sets on the base of the new sampling rate using interp1.m.
If enable is off a change of the Sampling rate value in the main edit window does not down- or upsample the data. Just the timing will be changed.
(If any data file is imported without any information about the sampling rate, this value will be set automatically to 1000. Change this value to the correct sampling rate if known.)
=> If the user samples first down and then up using the same factor the result will not be identical to that before the sampling as information is lost with down sampling.
The other way round the result should be the same as before
Calculate : GP/RMS
see Calculate :Potential
The only difference is that GP or RMS values are used.
Important: The user has to calculate GP/RMS first.
Calculate: Symmetry
Compute a measure of symmetry for every column of a given data matrix, when a list of
pairs of channels is provided.
Depending on "NormFlag", this measure is either the sum of differences of absolute values at corresponding sensors divided by norm of the data vector ('Abs.'), or the sum of differences of the values at the corresponding sensors, divided by the norm of the data vector ('Sign.'), or the sum of differences of the values at the corresponding sensors, each divided by the sum of the absolute values at the corresponding sensors ('Lat' => Lateralization Index) or the same as Lat but after a z-normalization of the data matrix (Norm. Lat.) or the same as (Norm. Lat.) but the sum of the absolute differences of the values at the corresponding sensors which always results in a positive symmetry index.
Program: CalcSymmetry.m
The resulting symmetry courses as the used pairs will be plotted in an extra window.
To export the symmetry data use Export: User Data
Get Pairs: The user has to open an ascii file with given pairs of the chosen sensors configuration. Look at Emegs2dUtil: SymmetryPairs :129LateralSymPairs as an example of 59 given pairs of the default 129 channel configuration to calculate a kind of lateralization between left and right hemispheres.
Set Pairs: If the pairs are not given the user could prepare the pairs in any text file (please use the upper example) or choose a number of pairs interactively from a special sensor configuration window . If the user does the Set Pairs menu he/she is asked for a file name in which the pairs will be stored.
Calculate: Grand Mean
Calculates the grand mean over all files the user has to choose and set it to the Actual file.
The user has to choose one file first to set the plot variables. If the plot of this file is completed the user will be asked for the second file. Choose this and continue with all files you want to add to the grand average. If you have chosen the last file stop adding with Cancel.
At this stage calculation of Grand Average is only available
with
AsciiBesaVec
AsciiHeaderVec
or SCADS file format.
Calculate: Rotate Sensor Config.
If a sensor configuration is already set(using File:Open Sensor File) it is possible to rotate the configuration around the x,y or z axis. To do this input the the required values in degree and press Apply. The rotation follows the right hand rule: Put the thumb of your right hand in the direction of the axis. (x=> from center to right ear, y=>from center to nose, z=>from center to vertex). The direction of the rotation corresponds to the curvature of the fingers of this hand.
Example: A z-rotation of -90 degrees rotates a nasion sensor towards the right ear. A positive rotation value leads to a counterclockwise rotation. (To save this electrode configuration and use it in the following processing press save. The new rotated configuration will be saved in SCADS format using the filemask Rot Nchan.sensor configuration. If you want to save it in a different sensor file format also please use Export:Sensor File. Important: As long as you have not pressed SaveEmegs2d will not use the new rotated configuration.
Calculate:
Reset
Sets all data back to the data, which was given before the calculation has been done. Reset files to previous ones
View:Negativ:Up or Down
Defines the view in the main window. .